In big science communities such as genomics, climate, and weather sensing, data lakes and ponds exist inside an ecosystem of centralized as well as individual ad-hoc repositories, each with different publication and data access standards. The genomics community, the primary science driver for this project, uses a wide range of metadata standards for publishing and searching experiment records. Repositories employ various data access protocols over TCP/IP point-to-point delivery, ranging from FTP to SFTP, HTTP(S), Aspera, Globus, as well as various application specific transfer procedures. For genomics datasets that are hosted on centralized repositories, such as the NCBI Sequence Read Archive (SRA), the EU/UK ENSEMBL database, or NASA’s GeneLab, they are generally well curated and replicated for availability. However only a fraction of all raw and processed genomics data are hosted in these repositories; another large portion is published by individual researchers and small research communities using whatever resources available to them. As a result, many datasets are organized in ad hoc ways, dependent on complex and manual mechanisms for access control, and not replicated, making their availability subject to hardware or network failures. While publishers spend significant time and resources to make their data available, the consumers are often unaware of the availability of these datasets and/or unable to utilize them.
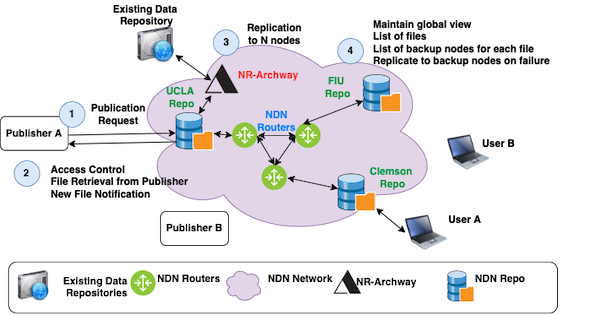
We propose to develop SDSF:Hydra - a secure, distributed storage framework – a framework that supports big-science communities with secure and resilient access to big datasets. Different from distributed databases, SDSF will be a loose federation of repositories potentially owned by multiple administrative entities between organizations, and will have security built into its file operations and data access natively. SDSF will address the following two problems at once: providing data publishers an automatically replicated, location independent publication platform for new data, and providing data consumers a uniform name-based interface to access all data, including both newly published data and existing data stored in legacy storage systems. SDSF will (a) provide long term data storage, (b) automatically replicate data across multiple geographically distributed repositories and maintain a desired degree of data replication in face of individual repository failures, (c) provide secure and scalable file access, and (d) provide a unified interface to access data stored in both SDSF and existing storage systems.
This project aims to improve the publication and accessibility of big science data starting with the genomics community. By addressing both research and engineering challenges, we aim to achieve the following deliverables: (i) leveraging the results from 10-year Named Data Networking (NDN) research, design and develop a distributed file repository software (NDN DF-Repo); (ii) leveraging the security solutions from NDN research, provide easy-to-use security and access control mechanism for the data publisher; these two deliverables will accomplish tasks (a)-(c) stated above; (iii) Designing and developing an NDN DF-Repo enhancement module NDR-Archway, which will add the ability for NDN DF-Repo to serve files from existing data publication systems, providing users a unified interface to access all data; and (iv) deploying SDSF over FABRIC as a pilot for simulated experimentation at scale. Note that SDSF is not a storage infrastructure but a software framework that the communities can deploy on their own infrastructure.
Intellectual Merit
By leveraging the results from 10-year NDN research, this work will create a distributed federation of file repositories that enables scientific communities to systematically store and access large amounts of data scalably and resiliently. The federation will be loosely coupled based on domain-relevant names and offer easy integration with existing scientific workflows. The federation will self-organize, automatically accommodating new repositories joining and maintaining a degree of file replication in face of node failures and recoveries.
Broader Impacts
Scalable and resilient access to all data is a basic requirement to support data-intensive science and engineering research. SDSF will enable secure and scalable data access to large numbers of users to alleviate the publication and data access problems facing genomics communities. Similar problems also exist in other scientific communities (e.g. climate, meteorology, astrophysics, and geology), which can benefit from SDSF as a replicated data storage solution with data authenticity and reduced network traffic as well as data access latency.